Getting Started with R
To get started with the labs, you will need to install R and other supporting applications. Links to download all the software is on the Software page at http://data606.net/software which is available on the Course Overview menu. Once you are in RStudio, you will need to install a number of R packages using the following two commands:
install.packages(c('openintro','OIdata','devtools','tidyverse', 'ggplot2',
'psych','reshape2','knitr','markdown','shiny','R.rsp',
'fivethirtyeight'))
devtools::install_github("jbryer/DATA606")Note that this course used to be listed as IS 606, hence the different package name. The library command will load the package and the startLab function will:
- Create a folder called
Lab1in the current working directory. - Open the the RMarkdown file (note that the name should have your login name).
library(DATA606)
startLab('Lab1')Note the Lab1 directory has been created.
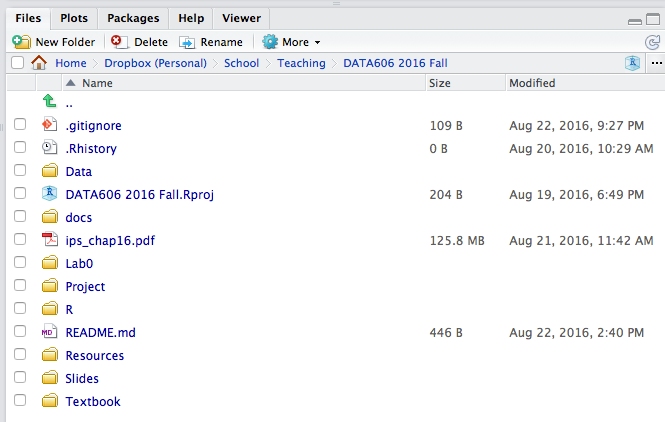
RStudio Files Panel
If you click Lab0 that will take you into that folder where you will see the RMardkown file (.Rmd extension) along with all the supporting files. Under the More menu there is an option to Set As Working Directory, you will want to do this before working in the RMarkdown file.
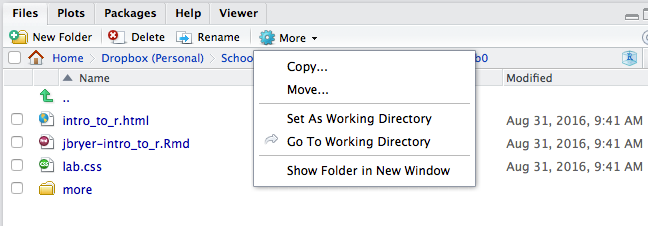
Setting Working Directory
The console pane will always tell you what your current working directory is. Clicking the arrow icon will open that folder in the Files tab on the right. R will look for files and directories that do not have absolute paths (i.e. starting with C: on windows or / on Linux and Macs) relative to the working directory.
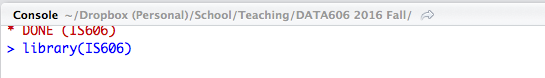
Working Directory